Case Study: Scaling Rare Disease Research in the Azure Cloud
When Fondation 101 Génomes was looking to build the world’s largest collection of Marfan Syndrome genomes to study this rare disease (and others), they turned to the Azure Cloud. With Tuple’s help, we built a genomics data lake that is now helping to scale their research.
With the support of the King Baudouin Foundation in Belgium, Ludivine and Romain Alderweireldt-Verboogen created Fondation 101 Génomes (F101G) to help children who, like their son, suffer from a rare disease known as Marfan Syndrome.

Marfan Syndrome is a connective tissue disorder that is primarily caused by mutations in the fibrillin-1 (FBN1) gene, though there are many mutations that are associated with this syndrome. In some patients, the syndrome simply causes elongated limbs and fingers. In others, it may lead to severe cardiovascular or pulmonary problems. However, the genetic reasons behind the diverse range in effects of this syndrome remain mostly unknown. It is believed that there are some protective genes (through epistatic interactions with FBN1) that are yet to be discovered and could be exploited therapeutically.

To aid in the research of Marfan Syndrome, F101G set out to create the world’s largest genome repository of data from affected patients. By combining large cohorts of Marfan genomes with accompanying clinical, phenotype, and radiologic imaging data, they hope to provide researchers with an environment in which they can better study and understand this condition.

Azure Architecture
The architecture for the solution is centered around the genomics data lake. Once data lands in the data lake from various input sources, we can organize, analyze, and visualize this data through services such as Azure Databricks and Azure Synapse Analytics.
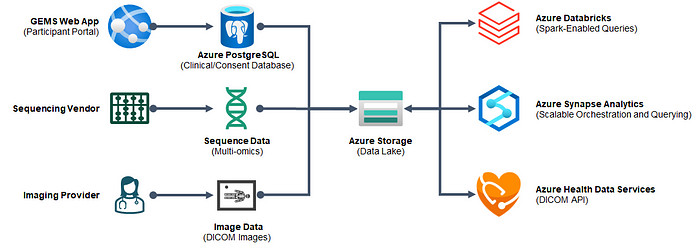
- Enrollment and consent data comes from F101G’s GEMS application (in Azure Web Apps)
- -omics data are loaded from a sequencing vendor (using a blend of Azure Databricks and Azure Synapse pipelines)
- Image data is loaded from external imaging providers (using Synapse pipelines)
Once the data are processed and organized in the data lake, we can then begin querying the data at-scale using Azure Databricks. Plus, we can also serve up the DICOM images to the appropriate viewers for annotation using the Azure Health Data Services API.
How Does the Cloud Help with this Research?
Data lakes are perfect for housing tons of heterogeneous patient data in one place. Not only are we needing to store the raw sequence data, but also the processed variant files, DICOM images of a patient’s MRI/CT scan, and phenotype information. This means that data lake organization is key to success when we want to start asking questions across the full dataset.

Because of the way we’ve organized the F101G genomics data lake, questions like these are a breeze to answer:
- “How many participants are homozygous for the G;G genotype for SNP rs147195031 in the FBN1 gene?”
- “Is there a correlation in any of the variants in the NOTCH3 gene and the various Marfan phenotypes?
- “Compared to the control population, do any genetic variants appear statistically more in our Marfan patients that may explain the cardiovascular effects of the syndrome?”
Using tools like Azure Databricks with the Glow package (for reading in VCF data in PySpark), we can query across the entire data lake of samples in seconds to minutes. This enables us to understand the genetic makeup of the Marfan genomes to help ascertain what might be at play in term of the variation in severity of the disease.

Plus, processing terabytes of genomes takes a considerable amount of compute power. However, it’s doesn’t necessarily mean that this processing has to take forever. With flexible cloud compute resources, we can scale to process hundreds to thousands of genomes through bioinformatics pipelines at once. This reduces the time to insight drastically by automating and distributing the workload on-demand.
So, What’s Next?
Our next steps include expanding out the computational analysis capabilities in the F101G cloud environment. F101G’s goal is to allow external collaborators to use a suite of tools to explore and analyze the multi-omics data, DICOM images, and clinical information, all through a secure, access-controlled framework.
In addition, F101G’s future goal is to expand into other rare diseases, which will continue to grow the genomics data lake in their Azure environment. Together, we’ll be exploring various cloud-native tools and capabilities to automate the bioinformatics processing of the data and develop revolutionary analytical and machine learning approaches to help solve the biological mysteries of Marfan syndrome and beyond.
Want to Tackle a Disease, Too?
The beauty of F101G and the cloud architecture that we’ve built is that it’s meant to be shared. If you’re interested in working on another disease, we invite you to join in on the F101G effort. You can collaborate with F101G and host the -omics data of another rare disease on F101G’s infrastructure or deploy a similar infrastructure on your own environment. F101G hopes to increase the statistical power of their research in Marfan syndrome and beyond by sharing in the collaborative fight against rare diseases using genomics and cloud computing.
If you’d like to work with us to build out your cloud capabilities and tackle another disease, please Contact Us today! Our team specializes in building cloud-based architectures to solve complex problems in genomics and beyond.
