Supporting the Fight Against Cancer, Tuple Unveils Anti-PD-1 Antibodies Fully Designed by AI
In a recent paper released in Technology in Cancer Research & Treatment, Tuple unveils its latest effort: designing antibodies using AI for the treatment of cancer.
In this work, researchers employed the EvoDiff system (from Microsoft Research) to perform protein diffusion. Protein diffusion works in a similar way to how ChatGPT generates text or how an image diffusion model can make a cool picture of a cat in a spacesuit in the style of van Gogh.

Protein diffusion in the design of antibodies is a new frontier with great promise in the field of drug discovery. Instead of starting with the lab-based process of selecting antibody candidates, we can provide the AI system with examples of other PD-1 targeting antibodies. Then, in short, the system will generate novel amino acid sequences that fold into parts of an antibody. With any luck, these generated antibodies behave similarly to naturally derived ones.
Using the highly scalable protein docking pipeline described in our last post (previously used in a SARS-CoV-2 modeling project with MIT Lincoln Lab), each of the generated antibodies was docked against PD-1, an important protein target in cancer therapies.
The in silico approach described in the preprint showcases the utility of AI in drug discovery and high-performance computing in the rapid assessment of any drug candidates that come out of the AI system.
PD-1 and It’s Role in Cancer
The programmed cell death protein 1 (PD-1, CD279) is an important therapeutic target in many oncological diseases. This checkpoint protein inhibits T lymphocytes from attacking other cells in the body. Thus, blocking this interaction improves the clearance of tumor cells by the immune system.

Anti-PD-1 antibodies block the interaction of PD-1 with it’s ligand(s), PD-L1 and PD-L2, and thus allows the T cell to perform an immune attack against the cancer cell.

While there are already multiple FDA-approved anti-PD-1 antibodies, including blockbusters like nivolumab (Opdivo from Bristol-Myers Squibb) and pembrolizumab (Keytruda from Merck), there is still a need to discover new and improved checkpoint inhibitor therapeutics.
The TUPPD1 Candidates
33 reference sequences of existing PD-1-targeting antibodies were collected from Thera-SAbDab, the “Therapeutic Structural Antibody Database” from the Oxford Protein Informatics Group. These reference sequences represented a broad swath of existing antibodies that target PD-1, some of which are used therapeutically today. These sequences were used to guide the diffusion process of the AI system — called “conditional diffusion”.
The conditional protein diffusion process generated 3 heavy chain sequences and 3 light chain sequences, which were then folded into the tertiary protein structures (using ColabFold) and combined to form 9 antibody candidates.
Note: The antibody structures tested in this study only contain the Fv portion of the immunoglobulin G proteins.
These 9 candidates, along with the 33 reference antibodies, were docked to PD-1 using HADDOCK, a flexible docking system for building biomolecular complexes. Then, binding affinity metrics were collected to assess the diffused proteins’ binding versus that of the reference antibodies.
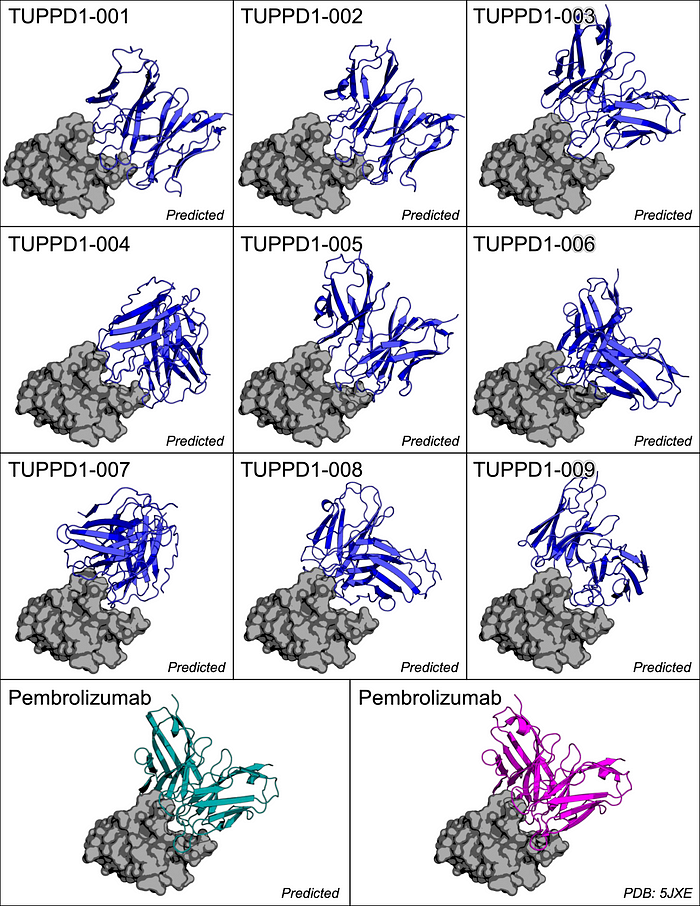
Across various metrics, some diffused antibodies performed worse than the reference set of antibodies. However, there are specific candidates (TUPPD1–001, 002, 009) that performed similarly to the reference antibodies.

What’s Left to Do?
This preliminary study serves a starting point for the development of novel therapeutics though a purely in silico yet scalable pipeline. Further enrichment of the reference dataset for the conditional diffusion should be performed to optimize the generation outputs and better mimic naturally derived antibodies. Then, additional antibodies will be generated and tested.
Pipeline Improvement
As shown in Tomezsko and Ford et al. 2023, large-scale protein docking can now be achieved through high-performance computing and pipeline containerization. This includes the automation of the experiment generation and post processing.
However, for antibody diffusion (or protein generation in general), there is a need to better automate the curation of reference sequences for the conditional diffusion process and to fold many antibody structures efficiently. These steps will be improved in Tuple’s future research efforts.
Onto the Lab!
This study is purely computational, but shows promise in a few of the candidate structures. Soon, the best candidates (TUPPD1–001, 002, 009) will be promoted to in vitro studies for empirical validation. This will help to build support and proof that AI-based antibody development is a viable approach to streamlining the otherwise difficult and expensive lab-based workflow.
Stay tuned for more AI-fueled bioinformatics and genomics research coming soon!
Resources:
- Paper: https://www.biorxiv.org/content/10.1101/2024.01.18.576323
- GitHub Repository: https://github.com/tuplexyz/PD-1_Fab_Diffusion
Stay Curious…
